Computational Simulations
This content is adapted from a workshop developed and presented by Dr. Priyanka Prakash, Research Computing Scientist here at UVA Research Computing.
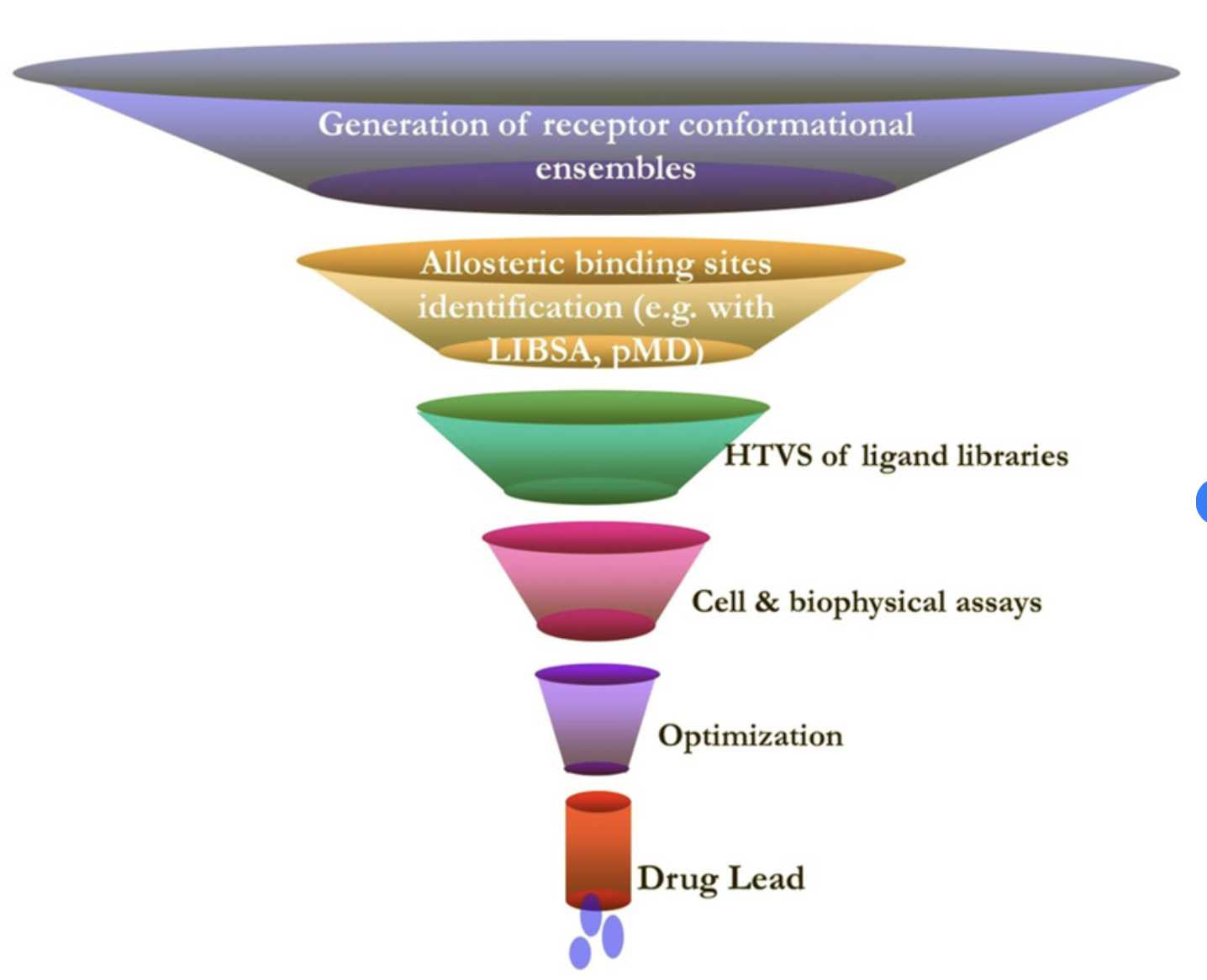
Dr. Prakash’s articles, which focus on small-molecule therapeutics, give examples of how computational methods are used when the protein structure is known but potential binding pockets are not. These tools can help identify where to target a drug, even when many similar proteins exist—enabling greater selectivity.
To explore the tools and research she referred to, you can find links to her publications on her LinkedIn page: https://www.linkedin.com/in/priyaps.
Simulations also play a role in developing membrane blockers, helping researchers understand how molecules interact with membrane-bound proteins.
The overall process follows a funnel-shaped workflow (as pictured below): starting with broad computational modeling and predictions, then narrowing down to experimental testing and lead compound development (not yet a full drug).