Chemprop Application, Part III
Screening the ZINC Library with Chemprop
Following the discovery of Halicin, the Chemprop model was scaled up to screen for additional antibiotic candidates. The researchers applied the model to a massive chemical space — the ZINC compound library, which contains over 107 million molecules.
Large-Scale In-Silico Screening
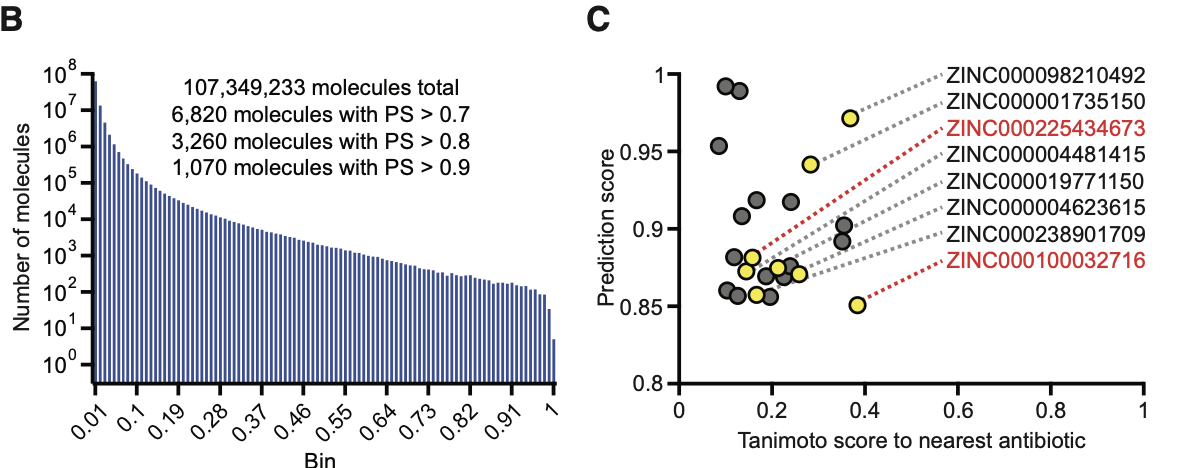
The figure below shows the results of this large-scale screen.
Out of the full set, the model predicted thousands of molecules to have high activity scores.
These were filtered based on prediction confidence.
Experimental Testing of Top Candidates
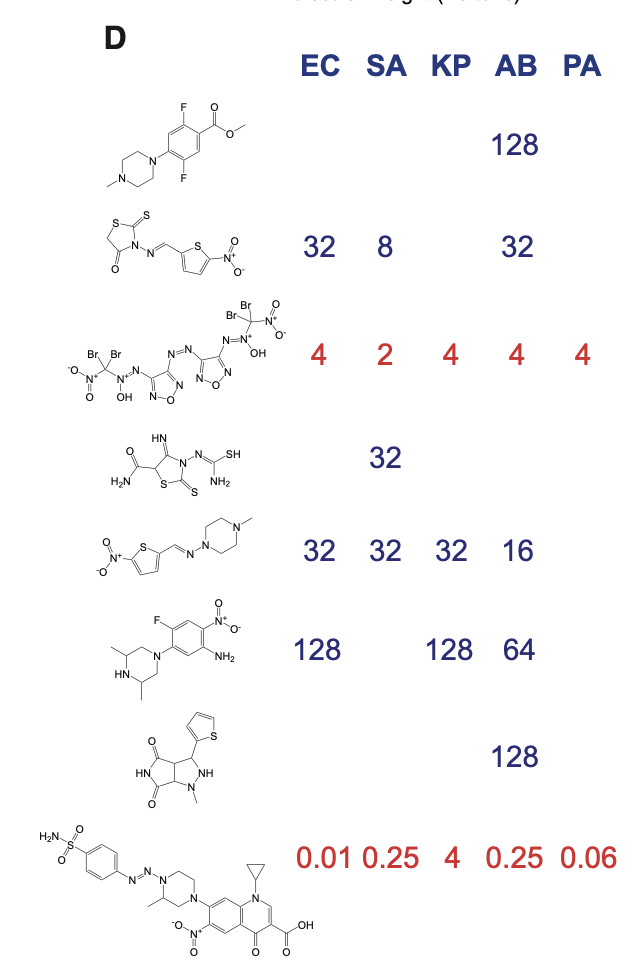
From this screen, several top candidates were selected for experimental testing.
Two of the compounds — ZINC000100032716 and ZINC000225434673 — showed strong promise in lab experiments.
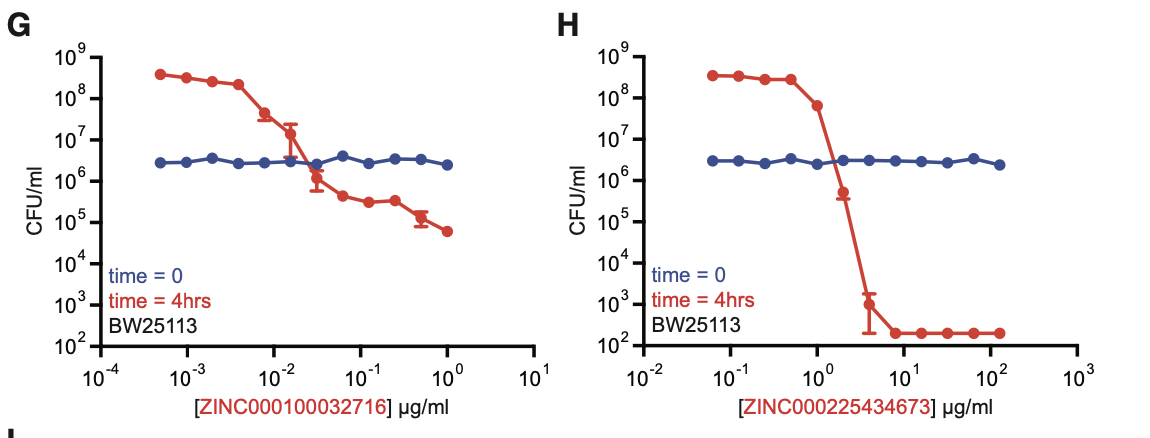
Chemprop’s in-silico screening helped identify potential new compounds with antibiotic-like properties.